COVID-19 Outbreak (Update: More than 2.9M cases and 132,313 deaths in US)
This is a really good paper, but I can't really follow a lot of it, since I'm not a doctor. Maybe someone here can explain a bit about the different mutations. I'm trying to understand if the "geo marker" (my term for how they know where it came from), and if one version is really any different than another? Does the ORF1a, ORF1b, S, etc mean anything?
https://www.ncbi.nlm.nih.gov/books/NBK554776/
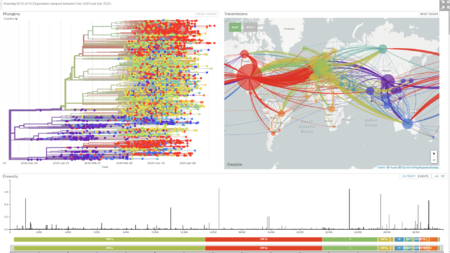
https://nextstrain.org/ncov
https://www.gisaid.org/
This is a really good paper, but I can't really follow a lot of it, since I'm not a doctor. Maybe someone here can explain a bit about the different mutations. I'm trying to understand if the "geo marker" (my term for how they know where it came from), and if one version is really any different than another? Does the ORF1a, ORF1b, S, etc mean anything?
https://www.ncbi.nlm.nih.gov/books/NBK554776/
Pathophysiology
CoVs are enveloped, positive-stranded RNA viruses with nucleocapsid. For addressing pathogenetic mechanisms of SARS-CoV-2, its viral structure, and genome must be considerations. In CoVs, the genomic structure is organized in a +ssRNA of approximately 30 kb in length — the largest known RNA viruses — and with a 5′-cap structure and 3′-poly-A tail. Starting from the viral RNA, the synthesis of polyprotein 1a/1ab (pp1a/pp1ab) in the host is realized. The transcription works through the replication-transcription complex (RCT) organized in double-membrane vesicles and via the synthesis of subgenomic RNAs (sgRNAs) sequences. Of note, transcription termination occurs at transcription regulatory sequences, located between the so-called open reading frames (ORFs) that work as templates for the production of subgenomic mRNAs. In the atypical CoV genome, at least six ORFs can be present. Among these, a frameshift between ORF1a and ORF1b guides the production of both pp1a and pp1ab polypeptides that are processed by virally encoded chymotrypsin-like protease (3CLpro) or main protease (Mpro), as well as one or two papain-like proteases for producing 16 non-structural proteins (nsps). Apart from ORF1a and ORF1b, other ORFs encode for structural proteins, including spike, membrane, envelope, and nucleocapsid proteins.[1] and accessory proteic chains. Different CoVs present special structural and accessory proteins translated by dedicated sgRNAs.
Pathophysiology and virulence mechanisms of CoVs, and therefore also of SARS-CoV-2 have links to the function of the nsps and structural proteins. For instance, research underlined that nsp is able to block the host innate immune response.[7] Among functions of structural proteins, the envelope has a crucial role in virus pathogenicity as it promotes viral assembly and release. However, many of these features (e.g., those of nsp 2, and 11) have not yet been described.
Among the structural elements of CoVs, there are the spike glycoproteins composed of two subunits (S1 and S2). Homotrimers of S proteins compose the spikes on the viral surface, guiding the link to host receptors.[8] Of note, in SARS-CoV-2, the S2 subunit — containing a fusion peptide, a transmembrane domain, and cytoplasmic domain — is highly conserved. Thus, it could be a target for antiviral (anti-S2) compounds. On the contrary, the spike receptor-binding domain presents only a 40% amino acid identity with other SARS-CoVs. Other structural elements on which research must necessarily focus are the ORF3b that has no homology with that of SARS-CoVs and a secreted protein (encoded by ORF8), which is structurally different from those of SARS-CoV.
In international gene banks such as GenBank, researchers have published several Sars-CoV-2 gene sequences. This gene mapping is of fundamental importance allowing researchers to trace the phylogenetic tree of the virus and, above all, the recognition of strains that differ according to the mutations. According to recent research, a spike mutation, which probably occurred in late November 2019, triggered jumping to humans. In particular, Angeletti et al. compared the Sars-Cov-2 gene sequence with that of Sars-CoV. They analyzed the transmembrane helical segments in the ORF1ab encoded 2 (nsp2) and nsp3 and found that position 723 presents a serine instead of a glycine residue, while the position 1010 is occupied by proline instead of isoleucine.[9] The matter of viral mutations is key for explaining potential disease relapses.
Research will be needed to determine the structural characteristics of SARS-COV-2 that underlie the pathogenetic mechanisms. Compared to SARS, for example, initial clinical data show less extra respiratory involvement, although due to the lack of extensive data, it is not possible to draw definitive clinical information.
The pathogenic mechanism that produces pneumonia seems to be particularly complex. Clinical and preclinical research will have to explain many aspects that underlie the particular clinical presentations of the disease. The data so far available seem to indicate that the viral infection is capable of producing an excessive immune reaction in the host. In some cases, a reaction takes place which as a whole is labeled a 'cytokine storm'. The effect is extensive tissue damage. The protagonist of this storm is interleukin 6 (IL-6). IL-6 is produced by activated leukocytes and acts on a large number of cells and tissues. It is able to promote the differentiation of B lymphocytes, promotes the growth of some categories of cells, and inhibits the growth of others. It also stimulates the production of acute phase proteins and plays an important role in thermoregulation, in bone maintenance and in the functionality of the central nervous system. Although the main role played by IL-6 is pro-inflammatory, it can also have anti-inflammatory effects. In turn, IL-6 increases during inflammatory diseases, infections, autoimmune disorders, cardiovascular diseases and some types of cancer. It is also implicated into the pathogenesis of the cytokine release syndrome (CRS) that is an acute systemic inflammatory syndrome characterized by fever and multiple organ dysfunction.
https://nextstrain.org/ncov
https://www.gisaid.org/
